Publishing Data
The LACMIP collection is a invaluable scientific resource, and strategic collection management helps to make it as accessible as possible. Digitizing specimen data is one way that we can increase accessibility. Published digital data can be used both as-is, and as a finding aid for researchers who may want to visit the collections in person. iDigBio and GBIF are the two primary portals we where share our collection-level and specimen-level data.
The Integrated Publishing Toolkit (IPT) is what we use to manage collection-level metadata. For example, this is the LACMIP resource on the IPT, which feeds this page on iDigBio plus this page on GBIF. This metadata lives in the IPT and can be edited there. The IPT also facilitates publishing specimen-level datasets via a CSV file and associated field mapping.
Set-up
The following is provided for general reference. If you are going through this process you may want to reach out to LACMIP staff and the Museum’s database manager for assistance.
IPT
The IPT is software for publishing both specimen data and collection-level metadata, as exemplified above. It can be installed on any server; NHMLA is currently using iDigBio’s installation of the IPT. The figure and text below provide a step-by-step explanation of what we use the IPT for.
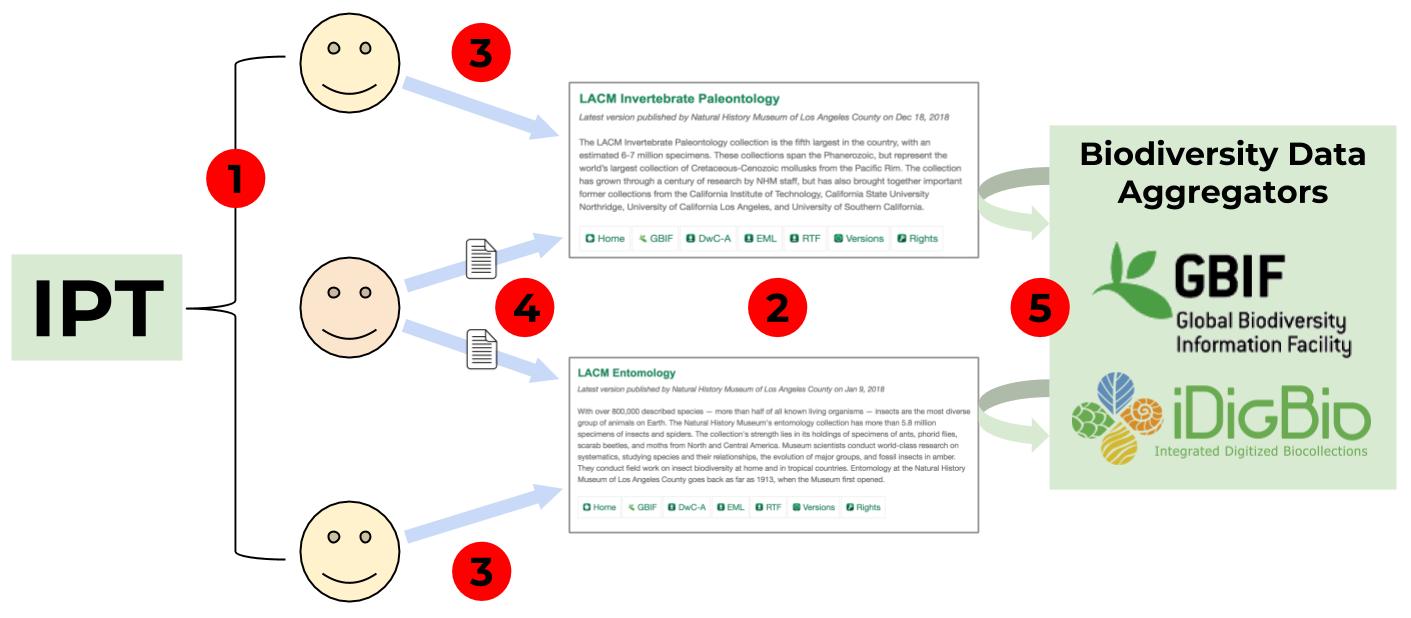
- Request user accounts on the iDigBio IPT for the NHMLA staff who need access, generally collections managers and informatics staff.
- Request an IPT “resource” for each collection. When requesting a resource you will need to provide your collection code, which combined with our institution code (LACM) forms a unique shortname, e.g. for Invertebrate Paleontology our collection code is “IP” and our shortname on the IPT is “lacm-ip.”
- Collections staff edit metadata fields directly on the IPT. Refer to the LACMIP resource on the IPT as an example.
- Informatics staff set up Darwin Core field mapping (see “Data in EMu” section below) and upload a CSV file for each collection dataset that is ready to publish.
- Biodiversity data aggregators harvest the CSV file and make the data available via their online portals.
Occurrence Data in EMu
In addition to setting up your collection resource on the IPT, you also need to determine what fields from EMu should be published. This is the most difficult part of publishing specimen data, but should only need to be done once (and modified when necessary). You will need to work closely with the Museum’s database manager to decide what EMu fields should get translated into which Darwin Core standard fields. For an example, see the LACMIP-EMu-to-DwC_[date].xlsx crosswalk (find it in Dropbox > EMu > IPT Data Publishing).
The crosswalk you develop will be coded into an EMu report, which can be run whenever you want to update your published specimen data. It is essential that when you are working on publishing your first dataset you check the report output very carefully to catch data quality issues, either with the way the report is set up or with your data itself.
You may not want every record in your EMu Catalogue module to be published. LACMIP excludes records by checking Publish on Internet = “No” on the Security tab of the Catalogue module. As part of your data verification process, make sure to check that any records which should not be published are indeed excluded.
Once the data being reported look accurate, the Museum’s database manager will upload this specimen occurrence file to the IPT, where iDigBio and GBIF harvest data from on a regular basis. Please note that it may be weeks to a month between when your data are uploaded to the IPT and when they become available on the aggregators. Uploading a new specimen occurrence file to the IPT will refresh your published data.
Media
If you have images or other media associated with specimens, these can (and should) also be shared via the IPT. Media metadata is shared in an Audubon Core file, which is uploaded to the IPT separately from your Darwin Core specimen occurrence data. Work with the Museum’s database manager to map fields from EMu to Audubon Core; for an example, see the LACMIP-EMu-to-AC_[date].xlsx crosswalk (find it in Dropbox > EMu > IPT Data Publishing).
Only derivative JPG images are uploaded to EMu for ease of reference by LACMIP staff. However, while the images in EMu are not directly shared online, the security settings for the images in EMu determine whether the corresponding images in the Museum’s DAMS (=Extensis Portfolio) are shared to aggregators.
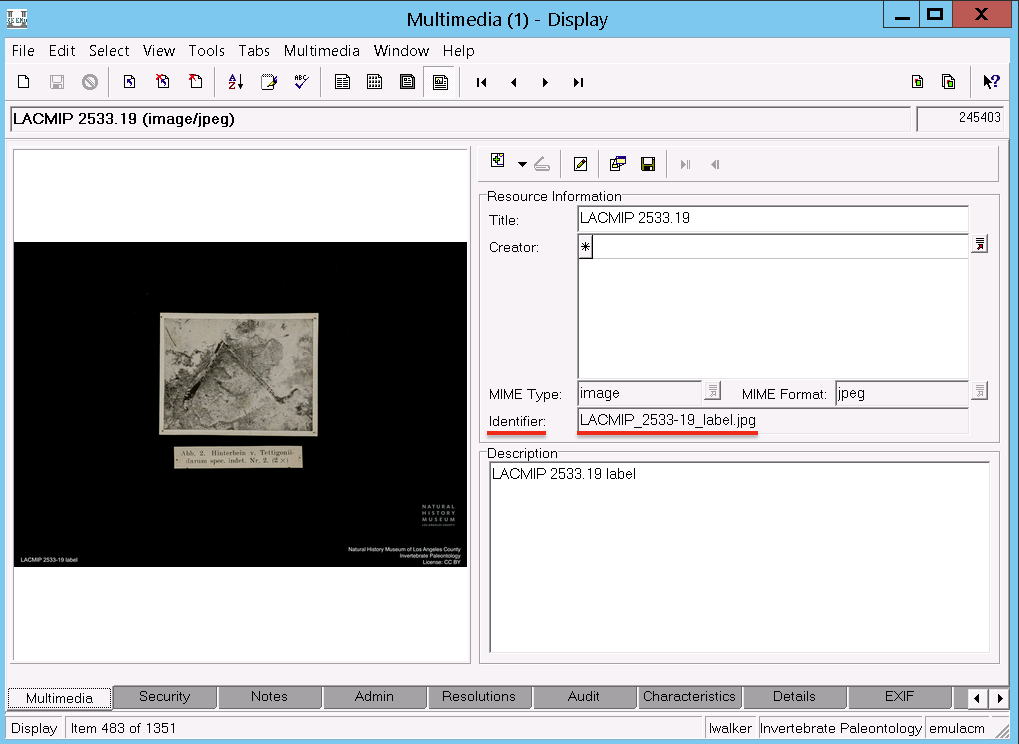
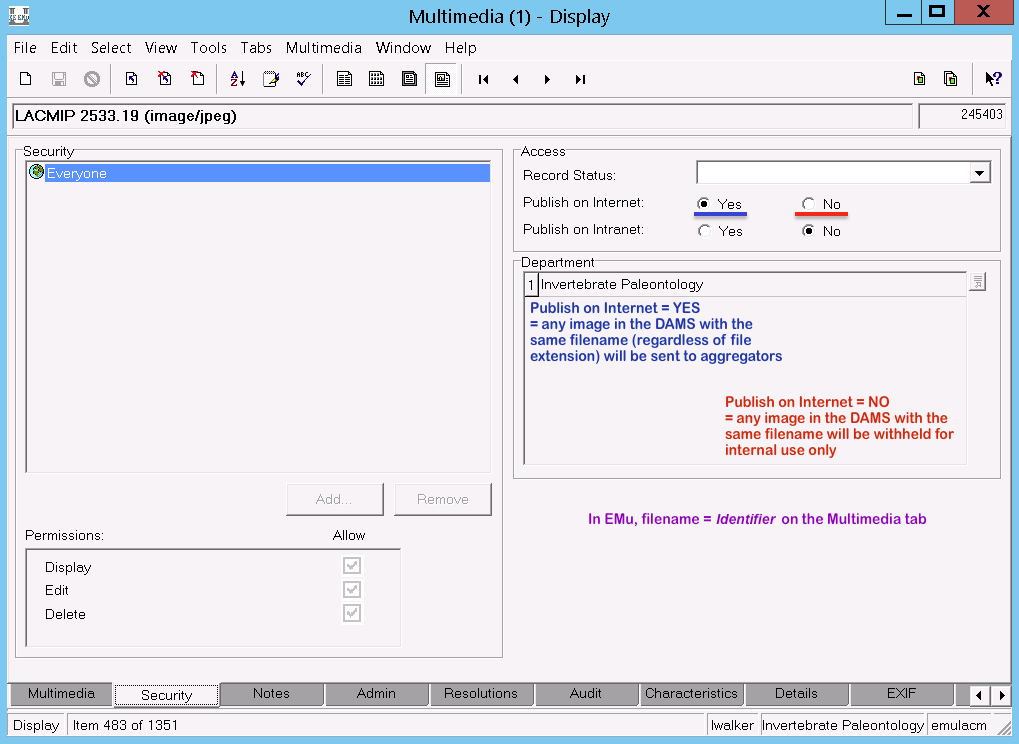
Images (archived and derivatives) are stored on an internal museum server. Permission to access these folders must be granted by the IT department and can only be done on-site, preferably using a tethered ethernet connection. Images can be added in bulk to LACMIP’s folders (IP Grant, IP Archive) by copying them straight into the appropriate folder on the server; however, all other management of these files (moving, renaming, etc.) must be done through the Extensis Portfolio interface.
Images saved to the “IP Grant” folder can be shared publicly; images saved to the “IP Archive” folder cannot be externally accessed. The latter folder primarily exists for archiving original, unedited images (DNGs, TIFs).
Maintenance
Remember to update the data and associated media that you have published based on whatever frequency makes sense for your collection, but probably at least annually. If you are actively digitizing or otherwise making changes/additions to your data in EMu you may want to update your IPT data more frequently.